PYCR1
| PYCR1 | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | PYCR1, ARCL2B, ARCL3B, P5C, P5CR, PIG45, PP222, PRO3, PYCR, pyrroline-5-carboxylate reductase 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 179035 MGI: 2384795 HomoloGene: 56002 GeneCards: PYCR1 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Pyrroline-5-carboxylate reductase 1, mitochondrial is an enzyme that in humans is encoded by the PYCR1 gene.[5][6]
This gene encodes an enzyme that catalyzes the NAD(P)H-dependent conversion of pyrroline-5-carboxylate to proline. This enzyme may also play a physiologic role in the generation of NADP(+) in some cell types. The protein forms a homopolymer and localizes to the mitochondrion. Alternate splicing results in two transcript variants encoding different isoforms.[6] As reported by Bruno Reversade and colleagues, PYCR1 deficiency in humans causes a progeroid disease known as De Barsy Syndrome mainly affecting connective tissues with dermis thinning and bone fragility.[7]
References
- ^ a b c GRCh38: Ensembl release 89: ENSG00000183010 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000025140 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Dougherty KM, Brandriss MC, Valle D (January 1992). "Cloning human pyrroline-5-carboxylate reductase cDNA by complementation in Saccharomyces cerevisiae". The Journal of Biological Chemistry. 267 (2): 871–5. doi:10.1016/S0021-9258(18)48364-0. PMID 1730675.
- ^ a b "Entrez Gene: PYCR1 pyrroline-5-carboxylate reductase 1".
- ^ Reversade B, Escande-Beillard N, Dimopoulou A, Fischer B, Chng SC, Li Y, et al. (September 2009). "Mutations in PYCR1 cause cutis laxa with progeroid features". Nature Genetics. 41 (9): 1016–21. doi:10.1038/ng.413. PMID 19648921. S2CID 10221927.
Further reading
- Herzfeld A, Legg MA, Greengard O (September 1978). "Human colon tumors: enzymic and histological characteristics". Cancer. 42 (3): 1280–3. doi:10.1002/1097-0142(197809)42:3<1280::AID-CNCR2820420337>3.0.CO;2-P. PMID 212173. S2CID 38290701.
- Merrill MJ, Yeh GC, Phang JM (June 1989). "Purified human erythrocyte pyrroline-5-carboxylate reductase. Preferential oxidation of NADPH". The Journal of Biological Chemistry. 264 (16): 9352–8. doi:10.1016/S0021-9258(18)60538-1. PMID 2722838.
- Yeh GC, Roth EF, Phang JM, Harris SC, Nagel RL, Rinaldi A (May 1984). "The effect of pyrroline-5-carboxylic acid on nucleotide metabolism in erythrocytes from normal and glucose-6-phosphate dehydrogenase-deficient subjects". The Journal of Biological Chemistry. 259 (9): 5454–8. doi:10.1016/S0021-9258(18)91032-X. PMID 6201483.
- Herzfeld A, Greengard O (November 1980). "Enzyme activities in human fetal and neoplastic tissues". Cancer. 46 (9): 2047–54. doi:10.1002/1097-0142(19801101)46:9<2047::AID-CNCR2820460924>3.0.CO;2-Q. PMID 6253048.
- Yeh GC, Harris SC, Phang JM (April 1981). "Pyrroline-5-carboxylate reductase in human erythrocytes". The Journal of Clinical Investigation. 67 (4): 1042–6. doi:10.1172/JCI110115. PMC 370662. PMID 6894153.
- Maruyama K, Sugano S (January 1994). "Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides". Gene. 138 (1–2): 171–4. doi:10.1016/0378-1119(94)90802-8. PMID 8125298.
- Basch JJ, Wickham ED, Farrell HM (August 1996). "Pyrroline-5-carboxylate reductase in lactating bovine mammary glands". Journal of Dairy Science. 79 (8): 1361–8. doi:10.3168/jds.S0022-0302(96)76493-7. PMID 8880459.
- Suzuki Y, Yoshitomo-Nakagawa K, Maruyama K, Suyama A, Sugano S (October 1997). "Construction and characterization of a full length-enriched and a 5'-end-enriched cDNA library". Gene. 200 (1–2): 149–56. doi:10.1016/S0378-1119(97)00411-3. PMID 9373149.
- Meng Z, Lou Z, Liu Z, Li M, Zhao X, Bartlam M, Rao Z (June 2006). "Crystal structure of human pyrroline-5-carboxylate reductase". Journal of Molecular Biology. 359 (5): 1364–77. doi:10.1016/j.jmb.2006.04.053. PMID 16730026.
External links
- PDBe-KB provides an overview of all the structure information available in the PDB for Human Pyrroline-5-carboxylate reductase 1, mitochondrial (PYCR1)
- v
- t
- e
-
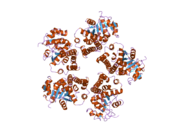 2ger: Crystal Structure and Oxidative Mechanism of Human Pyrroline-5-carboxylate Reductase
2ger: Crystal Structure and Oxidative Mechanism of Human Pyrroline-5-carboxylate Reductase -
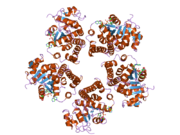 2gr9: Crystal structure of P5CR complexed with NADH
2gr9: Crystal structure of P5CR complexed with NADH -
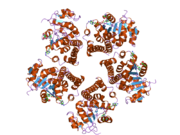 2gra: crystal structure of Human Pyrroline-5-carboxylate Reductase complexed with nadp
2gra: crystal structure of Human Pyrroline-5-carboxylate Reductase complexed with nadp -
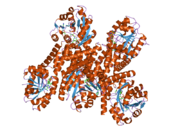 2izz: CRYSTAL STRUCTURE OF HUMAN PYRROLINE-5-CARBOXYLATE REDUCTASE
2izz: CRYSTAL STRUCTURE OF HUMAN PYRROLINE-5-CARBOXYLATE REDUCTASE
 | This article on a gene on human chromosome 17 is a stub. You can help Wikipedia by expanding it. |
- v
- t
- e