MMAB
| MMAB | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | MMAB, ATR, CFAP23, cblB, cob, methylmalonic aciduria (cobalamin deficiency) cblB type, metabolism of cobalamin associated B | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 607568 MGI: 1924947 HomoloGene: 12680 GeneCards: MMAB | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Cob(I)yrinic acid a,c-diamide adenosyltransferase, mitochondrial is an enzyme that in humans is encoded by the MMAB gene.[5][6][7]
Function
This gene encodes an enzyme (cob(I)yrinic acid a,c-diamide adenosyltransferase) that catalyzes the final step in the conversion of vitamin B12 into adenosylcobalamin (AdoCbl), a vitamin B12-containing coenzyme for methylmalonyl-CoA mutase.[7]
Clinical significance
Mutations in the gene are the cause of vitamin B12-dependent methylmalonic aciduria linked to the cblB complementation group.[7]
References
- ^ a b c GRCh38: Ensembl release 89: ENSG00000139428 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000029575 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Dobson CM, Wai T, Leclerc D, Kadir H, Narang M, Lerner-Ellis JP, et al. (December 2002). "Identification of the gene responsible for the cblB complementation group of vitamin B12-dependent methylmalonic aciduria". Human Molecular Genetics. 11 (26): 3361–3369. doi:10.1093/hmg/11.26.3361. PMID 12471062.
- ^ Leal NA, Park SD, Kima PE, Bobik TA (March 2003). "Identification of the human and bovine ATP:Cob(I)alamin adenosyltransferase cDNAs based on complementation of a bacterial mutant". The Journal of Biological Chemistry. 278 (11): 9227–9234. doi:10.1074/jbc.M212739200. PMID 12514191.
- ^ a b c "Entrez Gene: MMAB methylmalonic aciduria (cobalamin deficiency) cblB type".
External links
- GeneReviews/NCBI/NIH/UW entry on Methylmalonic Acidemia
- PDBe-KB provides an overview of all the structure information available in the PDB for Human Corrinoid adenosyltransferase (MMAB)
Further reading
- Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, et al. (February 2008). "Newly identified loci that influence lipid concentrations and risk of coronary artery disease". Nature Genetics. 40 (2): 161–169. doi:10.1038/ng.76. PMC 5206900. PMID 18193043.
- Hörster F, Baumgartner MR, Viardot C, Suormala T, Burgard P, Fowler B, et al. (August 2007). "Long-term outcome in methylmalonic acidurias is influenced by the underlying defect (mut0, mut-, cblA, cblB)". Pediatric Research. 62 (2): 225–230. doi:10.1203/PDR.0b013e3180a0325f. PMID 17597648.
- Keeratichamroen S, Cairns JR, Sawangareetrakul P, Liammongkolkul S, Champattanachai V, Srisomsap C, et al. (June 2007). "Novel mutations found in two genes of thai patients with isolated methylmalonic acidemia". Biochemical Genetics. 45 (5–6): 421–430. CiteSeerX 10.1.1.509.517. doi:10.1007/s10528-007-9085-y. PMID 17410422. S2CID 20799098.
- Kimura K, Wakamatsu A, Suzuki Y, Ota T, Nishikawa T, Yamashita R, et al. (January 2006). "Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes". Genome Research. 16 (1): 55–65. doi:10.1101/gr.4039406. PMC 1356129. PMID 16344560.
- Martínez MA, Rincón A, Desviat LR, Merinero B, Ugarte M, Pérez B (April 2005). "Genetic analysis of three genes causing isolated methylmalonic acidemia: identification of 21 novel allelic variants". Molecular Genetics and Metabolism. 84 (4): 317–325. doi:10.1016/j.ymgme.2004.11.011. PMID 15781192.
- Leal NA, Olteanu H, Banerjee R, Bobik TA (November 2004). "Human ATP:Cob(I)alamin adenosyltransferase and its interaction with methionine synthase reductase". The Journal of Biological Chemistry. 279 (46): 47536–47542. doi:10.1074/jbc.M405449200. PMID 15347655.
- Robertson NG, Khetarpal U, Gutiérrez-Espeleta GA, Bieber FR, Morton CC (September 1994). "Isolation of novel and known genes from a human fetal cochlear cDNA library using subtractive hybridization and differential screening". Genomics. 23 (1): 42–50. doi:10.1006/geno.1994.1457. PMID 7829101.
- v
- t
- e
-
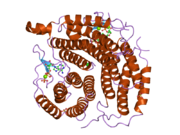 2idx: Structure of Human ATP:Cobalamin adenosyltransferase bound to ATP.
2idx: Structure of Human ATP:Cobalamin adenosyltransferase bound to ATP.
 | This article on a gene on human chromosome 12 is a stub. You can help Wikipedia by expanding it. |
- v
- t
- e